清华大学:《分子生物学》课程PPT教学课件(基因ene)第二十二章 核的剪切(Nuclear splicing)

Chapter 22 Nuclear splicing 清革大当
Chapter 22 Nuclear splicing

22.1 Introduction 22.2 Nuclear splice junctions are short sequences 22.3 Splice junctions are read in pairs 22.4 Nuclear splicing proceeds through a lariat 22.5 snRNAs are required for splicing 22.6 Ul snRNP initiates splicing 22.7 The E complex can be formed in alternative ways 22.8 5 snRNPs form the spliceosome 22.9 An alternative splicing apparatus uses different snRNPs 22.10 Group II introns autosplice via lariat formation 22.11 Alternative splicing involves differential use of splice junctions 22.12 cis-splicing and trans-splicing reactions 22.13 Yeast tRNA splicing involves cutting and rejoining 22.14 The unfolded protein response is related to tRNA splicing 22.15 The 3 ends of poll and polllI transcripts are generated by termination 22.16 The 3c ends of mRNAs are generated by cleavage 22.17 Cleavage of the 3c end may require a small RNA 22.18 Production of rRNA requires cleavage and modification events 22.19 Small RNAs are required for rRNA processing 清菜大当
22.1 Introduction 22.2 Nuclear splice junctions are short sequences 22.3 Splice junctions are read in pairs 22.4 Nuclear splicing proceeds through a lariat 22.5 snRNAs are required for splicing 22.6 U1 snRNP initiates splicing 22.7 The E complex can be formed in alternative ways 22.8 5 snRNPs form the spliceosome 22.9 An alternative splicing apparatus uses different snRNPs 22.10 Group II introns autosplice via lariat formation 22.11 Alternative splicing involves differential use of splice junctions 22.12 cis-splicing and trans-splicing reactions 22.13 Yeast tRNA splicing involves cutting and rejoining 22.14 The unfolded protein response is related to tRNA splicing 22.15 The 3¢ ends of polI and polIII transcripts are generated by termination 22.16 The 3¢ ends of mRNAs are generated by cleavage 22.17 Cleavage of the 3¢ end may require a small RNA 22.18 Production of rRNA requires cleavage and modification events 22.19 Small RNAs are required for rRNA processing

21.1 Introduction RNA splicing is the process of excising the sequences in RNA that correspond to introns,so that the sequences corresponding to exons are connected into a continuous mRNA. pre-mRNA heterogeneous nuclear RNA (hnRNA). hnRNP 清苇大当
RNA splicing is the process of excising the sequences in RNA that correspond to introns, so that the sequences corresponding to exons are connected into a continuous mRNA. pre-mRNA heterogeneous nuclear RNA (hnRNA). hnRNP 21.1 Introduction
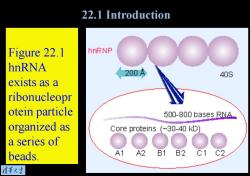
22.1 Introduction Figure 22.1 hnRNP hnRNA 200A 40S exists as a ribonucleopr otein particle 500-800 bases RNA_ organized as Core proteins (~30-40 kD) a series of beads. A1 A2 B1 B2 C1 C2 清菜大当
Figure 22.1 hnRNA exists as a ribonucleopr otein particle organized as a series of beads. 22.1 Introduction
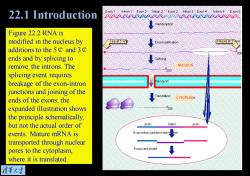
22.1 Introduction 4o2 Figure 22.2 RNA is modified in the nucleus by Cap at 5 en PolyA at 3 end additions to the5¢and3¢ ends and by splicing to remove the introns.The NUCLEUS 200 splicing event requires breakage of the exon-intron junctions and joining of the ends of the exons;the Transation CYTOPLASM expanded illustration shows 200 the principle schematically, but not the actual order of exon intron exon events.Mature mRNA is Exon-intron junctions are b ken transported through nuclear pores to the cytoplasm, Exons are joined where it is translated. 清菜大兰
Figure 22.2 RNA is modified in the nucleus by additions to the 5¢ and 3¢ ends and by splicing to remove the introns. The splicing event requires breakage of the exon-intron junctions and joining of the ends of the exons; the expanded illustration shows the principle schematically, but not the actual order of events. Mature mRNA is transported through nuclear pores to the cytoplasm, where it is translated. 22.1 Introduction

22.2 Nuclear splice junctions are interchangeable but are read in pairs GT-AG rule describes the presence of these constant dinucleotides at the first two and last two positions of introns of nuclear genes. Splice sites are the sequences immediately surrounding the exon-intron boundaries. 情菜大兰
GT-AG rule describes the presence of these constant dinucleotides at the first two and last two positions of introns of nuclear genes. Splice sites are the sequences immediately surrounding the exon-intron boundaries. 22.2 Nuclear splice junctions are interchangeable but are read in pairs

22.2 Nuclear splice junctions are interchangeable but are read in pairs Left (5)site Right(3)site Ex64G73G10062463 12Py N C6A1010N Exon Intron Figure 22.3 The ends of nuclear introns are defined by the GT-AG rule 清苇大当
Figure 22.3 The ends of nuclear introns are defined by the GT-AG rule 22.2 Nuclear splice junctions are interchangeable but are read in pairs

22.2 Nuclear splice junctions are interchangeable but are read in pairs Comrect splicing removes 3 introns by pairwise recognition ofthe junctions GU GU GU Figure 22.4 Splicing junctions are P airing of wrong junctions would remove exons as well as introns,but aberant products are not found recognized GU GU only in the correct pairwise GU GU rGU combinations. 清菜大当
Figure 22.4 Splicing junctions are recognized only in the correct pairwise combinations. 22.2 Nuclear splice junctions are interchangeable but are read in pairs
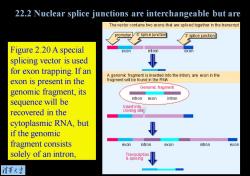
22.2 Nuclear splice junctions are interchangeable but are Thevector contains two exons that are spliced together in the transcript promoter5'splice junction 3splice junction Figure 2.20 A special exon int ron exon splicing vector is used for exon trapping.If an A genomic fragment is inserted into the intron;any exon in the exon is present in the fragment will be found in the RNA Genomic fragment genomic fragment,its intron exon intron sequence will be Insert into recovered in the cloning site cytoplasmic RNA,but if the genomic fragment consists exon intron exon intron exon solely of an intron, Transcription splicing 清第大当
Figure 2.20 A special splicing vector is used for exon trapping. If an exon is present in the genomic fragment, its sequence will be recovered in the cytoplasmic RNA, but if the genomic fragment consists solely of an intron, 22.2 Nuclear splice junctions are interchangeable but are read in pairs

Gene 5.6 kb 22.2 Nuclear splice junctions 11121314■516171 are interchangeable but are read in pairs mRNA=1.1 kb Figure 22.5 Northern Primary transcript(5.5 kb) blotting of nuclear RNA with an ovomucoid probe identifies discrete Lacks introns 5 6 precursors to mRNA.The Lacks introns 4,5,6,&7 contents of the more prominent bands are Contains only intron 3 indicated.Photograph kindly provided by Bert O'Malley. mRNA (1.1 kb) 清菜大当
Figure 22.5 Northern blotting of nuclear RNA with an ovomucoid probe identifies discrete precursors to mRNA. The contents of the more prominent bands are indicated. Photograph kindly provided by Bert O'Malley. 22.2 Nuclear splice junctions are interchangeable but are read in pairs
按次数下载不扣除下载券;
注册用户24小时内重复下载只扣除一次;
顺序:VIP每日次数-->可用次数-->下载券;
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第二十一章 转录的调控(Regulation of Transcription).ppt
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第二十章 转录的起始(Initiation of transcription).ppt
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第十九章 核小体(Nucleosomes).ppt
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第十八章 染色体(Chromosomes).ppt
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第十七章 DNA的重新排列(Rearrangement of DNA).ppt
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第十六章 逆转录病毒和逆转座子(Retroviruses and retroposons).ppt
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第十五章 转座子(Transposons).ppt
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第十四章 重组和修复(Recombination and repair).ppt
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第十三章 DNA复制(DNA replication).ppt
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第十二章 复制子(The replicon).ppt
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第十一章 噬菌体的战略(Phage strategies).ppt
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第十章 操纵子(The operon).ppt
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第九章 转录(Transcription).ppt
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第八章 蛋白质定位(Protein localization).ppt
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第七章 遗传密码的利用(Using the genetic code).ppt
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第六章 蛋白质合成(Protein synthesis).ppt
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第五章 信使RNA(Messenger RNA).ppt
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第四章 簇和重复(Clusters and repeats).ppt
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第三章 有多少基因(How many genes are there).ppt
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第二章 从基因到基因组(From Genes to Genomes).ppt
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第二十三章 催化RNA(Catalytic RNA).ppt
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第二十四章 免疫多样性(Immune diversity).ppt
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第二十五章 蛋白质间的开放交通(Protein trafficking).ppt
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第二十六章 信号的传输(Signal transduction).ppt
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第二十七章 细胞循环和生长调控(Cell cycle and growth regulation).ppt
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第二十八章 致癌基因和癌症(Oncogenes and cancer).ppt
- 清华大学:《分子生物学》课程PPT教学课件(基因ene)第二十九章 梯度、级联和发信号的途径(Gradients, cascades, and signaling pathways).ppt
- 四川大学:《植物生物学》课程教学资源(教案讲义)第一章 植物的细胞和组织.pdf
- 四川大学:《植物生物学》课程教学资源(教案讲义)第二章 植物体的形态结构和发育.pdf
- 四川大学:《植物生物学》课程教学资源(教案讲义)第三章 植物的无机营养.pdf
- 四川大学:《植物生物学》课程教学资源(教案讲义)第四章 光合作用.pdf
- 四川大学:《植物生物学》课程教学资源(教案讲义)第五章 植物的繁殖.pdf
- 四川大学:《植物生物学》课程教学资源(教案讲义)第六章 植物的生长发育及其调控.pdf
- 四川大学:《植物生物学》课程教学资源(教案讲义)第七章 生物多样性和植物的分类及命名.pdf
- 四川大学:《植物生物学》课程教学资源(教案讲义)第八章 藻类植物.pdf
- 四川大学:《植物生物学》课程教学资源(教案讲义)第九章 苔藓植物.pdf
- 四川大学:《植物生物学》课程教学资源(教案讲义)第十章 蕨类植物.pdf
- 四川大学:《植物生物学》课程教学资源(教案讲义)第十一章 裸子植物.pdf
- 四川大学:《植物生物学》课程教学资源(教案讲义)第十二章 被子植物.pdf
- 《环境工程微生物学》课程教学资源(PPT课件)第一章 细菌结构与形态(1/2).ppt
